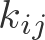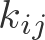$
\newcommand{\avg}[1]{\langle #1 \rangle}
\newcommand{\cc}[1]{[\mathrm{#1}]^{\mathrm{cell}}}
\newcommand{\cgdp}{\mathrm{C \! \cdot \! GDP}}
\newcommand{\cgtp}{\mathrm{C \! \cdot \! GTP}}
\newcommand{\comb}[1]{{#1}^{\mathrm{comb}}}
\newcommand{\conc}[1]{[\mathrm{#1}]}
\newcommand{\conceq}[1]{[\mathrm{#1}]^{\mathrm{eq}}}
\newcommand{\concss}[1]{[\mathrm{#1}]^{\mathrm{ss}}}
\newcommand{\conctot}[1]{[\mathrm{#1}]_{\mathrm{tot}}}
\newcommand{\cu}{\conc{U}}
\newcommand{\dee}{\partial}
\newcommand{\dgbind}{\Delta G_0^{\mathrm{bind}}}
\newcommand{\dgdp}{\mathrm{D \! \cdot \! GDP}}
\newcommand{\dgtp}{\mathrm{D \! \cdot \! GTP}}
\newcommand{\dmu}{\Delta \mu}
\newcommand{\dphi}{\Delta \Phi}
\newcommand{\dplus}[1]{\mbox{#1}^{++}}
\newcommand{\eq}[1]{{#1}^{\mathrm{eq}}}
\newcommand{\fidl}{F^{\mathrm{idl}}}
\newcommand{\idl}[1]{{#1}^{\mathrm{idl}}}
\newcommand{\inn}[1]{{#1}_{\mathrm{in}}}
\newcommand{\ka}{k_a}
\newcommand{\kcat}{k_{\mathrm{cat}}}
\newcommand{\kf}{k_f}
\newcommand{\kfc}{k_{fc}}
\newcommand{\kftot}{k_f^{\mathrm{tot}}}
\newcommand{\kd}{K_{\mathrm{d}}}
\newcommand{\kdt}{k_{\mathrm{dt}}}
\newcommand{\kdtsol}{k^{\mathrm{sol}}_{\mathrm{dt}}}
\newcommand{\kgtp}{K_{\mathrm{GTP}}}
\newcommand{\kij}{k_{ij}}
\newcommand{\kji}{k_{ji}}
\newcommand{\kkeq}{K^{\mathrm{eq}}}
\newcommand{\kmmon}{\kon^{\mathrm{ES}}}
\newcommand{\kmmoff}{\koff^{\mathrm{ES}}}
\newcommand{\kconf}{k_{\mathrm{conf}}}
\newcommand{\konf}{k^{\mathrm{on}}_{\mathrm{F}}}
\newcommand{\koff}{k_{\mathrm{off}}}
\newcommand{\kofff}{k^{\mathrm{off}}_{\mathrm{F}}}
\newcommand{\konu}{k^{\mathrm{on}}_{\mathrm{U}}}
\newcommand{\koffu}{k^{\mathrm{off}}_{\mathrm{U}}}
\newcommand{\kon}{k_{\mathrm{on}}}
\newcommand{\kr}{k_r}
\newcommand{\ks}{k_s}
\newcommand{\ku}{k_u}
\newcommand{\kuc}{k_{uc}}
\newcommand{\kutot}{k_u^{\mathrm{tot}}}
\newcommand{\ktd}{k_{\mathrm{td}}}
\newcommand{\ktdsol}{k^{\mathrm{sol}}_{\mathrm{td}}}
\newcommand{\minus}[1]{\mbox{#1}^{-}}
\newcommand{\na}{N_A}
\newcommand{\nai}{N_A^i}
\newcommand{\nao}{N_A^o}
\newcommand{\nb}{N_B}
\newcommand{\nbi}{N_B^i}
\newcommand{\nbo}{N_B^o}
\newcommand{\nc}{N_{C}}
\newcommand{\nl}{N_L}
\newcommand{\nltot}{N_L^{\mathrm{tot}}}
\newcommand{\nr}{N_R}
\newcommand{\nrl}{N_{RL}}
\newcommand{\nrtot}{N_R^{\mathrm{tot}}}
\newcommand{\out}[1]{{#1}_{\mathrm{out}}}
\newcommand{\plus}[1]{\mbox{#1}^{+}}
\newcommand{\rall}{\mathbf{r}^N}
\newcommand{\rn}[1]{\mathrm{r}^N_{#1}}
\newcommand{\rdotc}{R \!\! \cdot \! C}
\newcommand{\rstarc}{R^* \! \! \cdot \! C}
\newcommand{\rstard}{R^* \! \! \cdot \! D}
\newcommand{\rstarx}{R^* \! \! \cdot \! X}
\newcommand{\ss}{\mathrm{SS}}
\newcommand{\totsub}[1]{{#1}_{\mathrm{tot}}}
\newcommand{\totsup}[1]{{#1}^{\mathrm{tot}}}
\newcommand{\ztot}{Z^{\mathrm{tot}}}
% Rate notation: o = 1; w = two; r = three; f = four
\newcommand{\aow}{\alpha_{f}}
\newcommand{\awo}{\alpha_{u}}
\newcommand{\kow}{\kf} % {\kf(12)}
\newcommand{\kwo}{\ku} % {\ku(21)}
\newcommand{\kor}{\conc{C} \, \konu} % \konu(13)}
\newcommand{\kwf}{\conc{C} \, \konf} % \konf(24)}
\newcommand{\kro}{\koffu} % {\koffu(31)}
\newcommand{\kfw}{\kofff} % {\kofff(42)}
\newcommand{\krf}{\kfc} % {\kfc(34)}
\newcommand{\kfr}{\kuc} % {\kuc(43)}
\newcommand{\denom}{ \krf \, \kfw + \kro \, \kfw + \kro \, \kfr }
$